
Regulation of gene expression forms the foundation for basic cellular processes, development and disease in multicellular organisms. During my postdoctoral work in Vienna, I discovered that animal genomes, through rapid ‘arms race’ evolution, evolve new molecular mechanisms that allow the cell to bend and bypass the textbook rules of gene expression. Such ‘hacked’ gene expression produces RNA molecules that are essential for animals to combat genetic parasites such as transposons (‘jumping genes’). With my AIAS fellow project, I will build on these new concepts to identify the molecular mechanisms underlying ‘hacked’ genome expression, 2) investigate the connection between transposon silencing and general regulation of the genome. To address these questions, I will combine genetic manipulation of both in vivo models (Drosophila) and cell culture systems with molecular analysis using diverse genomics approaches. With my research as AIAS fellow I aim to pioneer the emerging field of heterochromatin-based expression mechanisms and uncover new modes of genome regulation during animal development and disease.
Project title
Innovation of genome regulation through arms race evolution in animals
Area of research
Molecular innovation in genetic conflicts
Fellowship period
1 Oct 2019 - 18 Nov 2022
Fellowship type:
AIAS-COFUND II Marie Skłodowska-Curie fellow

This fellowship has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 754513 and The Aarhus University Research Foundation.

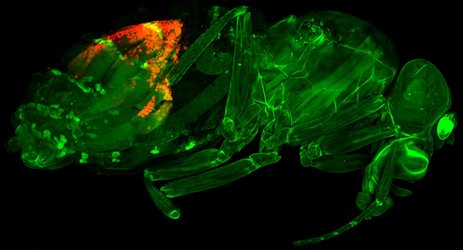
”A fruit fly (Drosophila melanogaster) with a recently evolved gene expression regulator that inhibits transposons (selfish jumping genes) stained in red. Image: Peter Refsing Andersen and Daniel Reumann”