Development of improved methods for predicting complex traits
Former fellow Doug Speed, together with colleagues from BiRC and NCRR, have developed improved methods for predicting complex traits. The work has been published in the journal Nature Communications this week.
Currently there is great interest in being able to use an individual's genetic information to predict their phenotypes, which are an individual's traits such as height, eye color and blood type. This is especially important for personalized medicine, which aims to accurately predict which individuals will develop particular diseases or will benefit from particular medications.
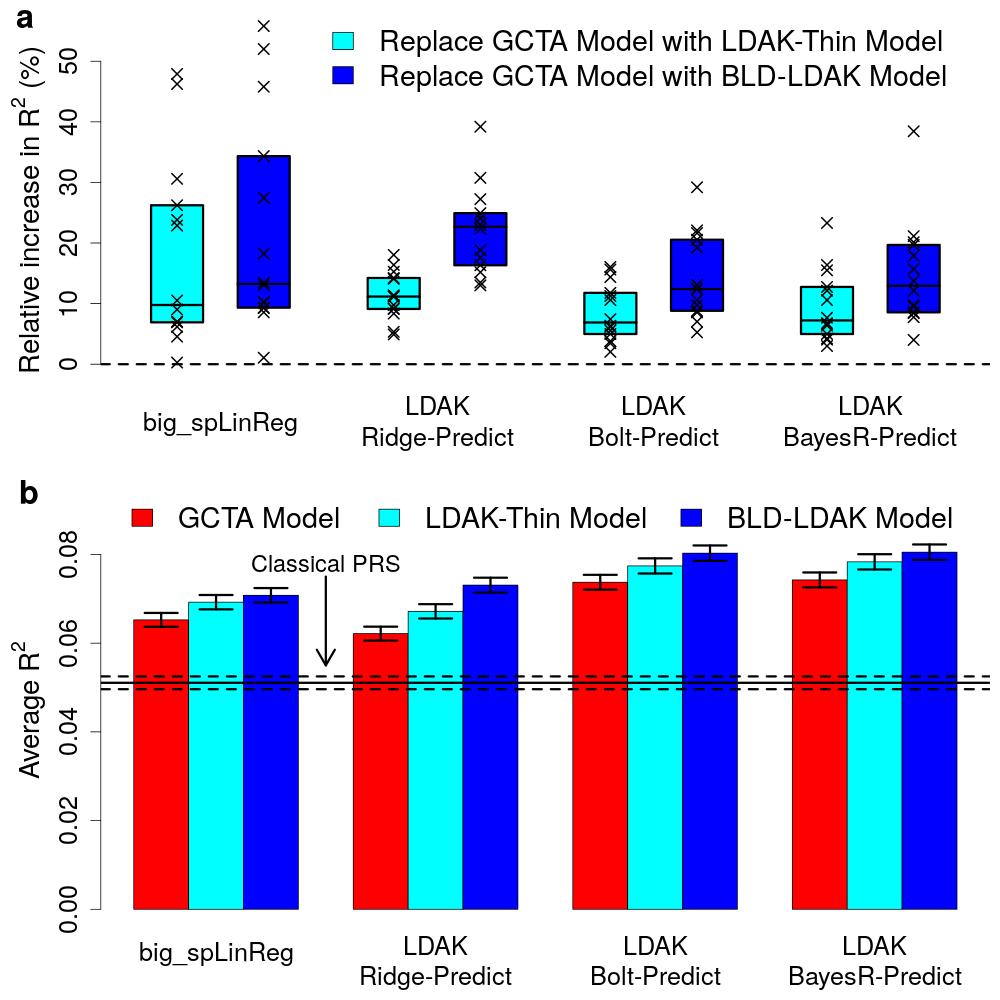
Doug Speed and colleagues have observed that most existing prediction tools assume that each genetic variant is equally important. This assumption is sub-optimal, because recent work has shown that the importance of a variant depends on factors such as its frequency, local levels of linkage disequilibrium and functional annotations. Therefore, this new paper presents eight new prediction tools that allow for alternative assumptions, and shows that this enables substantially improved prediction across a wide range of traits.
Four of the new tools use individual-level data. The paper shows that the best of these, LDAK-Bolt-Predict, outperforms the existing tools Lasso, BLUP, Bolt-LMM and BayesR for all 14 phenotypes considered. The remaining four new tools use summary statistics. The paper shows that the best of these, LDAK-BayesR-SS, outperforms the existing tools lassosum, sBLUP, LDpred and SBayesR for 223 of the 225 phenotypes considered. On average, the new tools outperform the existing tools by 14% (sd 1), which is equivalent to increasing the sample size by about a quarter.
"For personalized medicine to become a reality, we require models that can accurately predict an individual's phenotypes based on their genetic information. This work provides statistical tools for creating genetic prediction models that are substantially more accurate than existing tools," Doug Speed explained and continued:
"This work will have immediate benefit, as it means we can now better identify individuals who have high risk of developing different diseases."
The scientific article
You can read more about the new tools in the paper "Improved genetic prediction of complex traits from individual-level data or summary statistics", and try out the new tools in the software packages LDAK (www.ldak.org) and bigstatsr (https://github.com/privefl/bigstatsr).
Contact
Doug Speed, Professor, AIAS Former Fellow
doug@qgg.au.dk
Center for Quantitative Genetics and Genomics,
Aarhus University